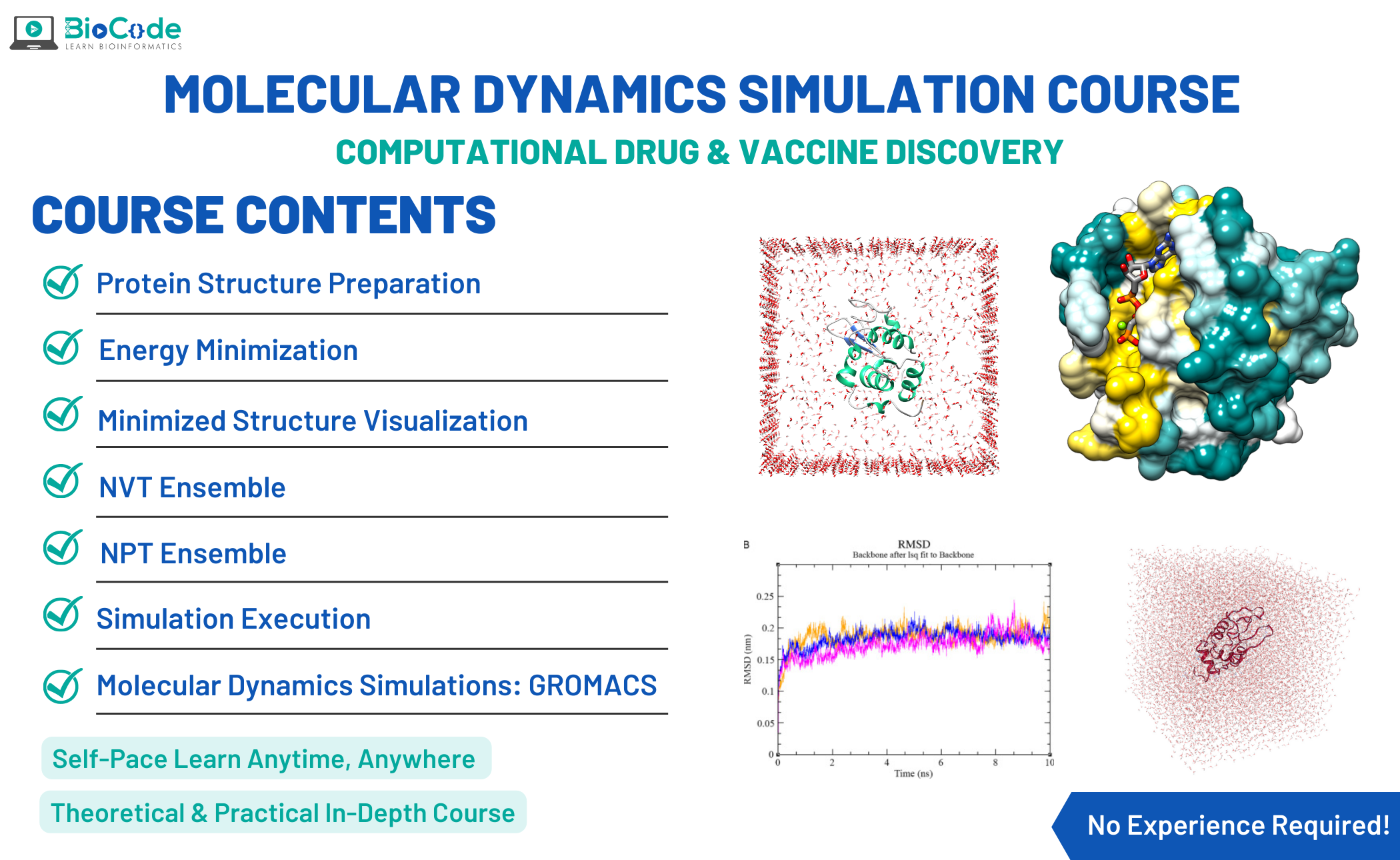
Molecular Dynamics Course
Molecular Dynamics Course - This course will focus on the theoretical underpinnings of md simulations, statistical mechanics principles, and practical tools and techniques for setting up and computing macroscopic. Up to 10% cash back the easiest way to learn molecular dynamics! Eukaryotic protein traffic and related topics, including molecular motors and cytoskeletal dynamics, organelle architecture and biogenesis, protein translocation and sorting,. Thanks to supercomputers, molecular dynamics (md) makes it possible to observe these processes with great precision and provides new knowledge of interest both in basic. Up to 10% cash back in this course, you will learn. (2) theory of molecular dynamics simulation. Describe what a force field is and what its. Central themes are the mechanisms and dynamics by which molecular structure and function evolve, how protein/ rna architecture shapes evolutionary trajectories, and how patterns in. Read reviews to decide if a class. Namd, recipient of a 2002 gordon bell award, a 2012 sidney fernbach award, and a 2020 gordon bell prize, is a parallel molecular dynamics code designed for high. Analysis of ekin, t (free dynamics) the atomic velocities of a protein establish a thermometer, but is it accurate? Explain the theory and mathematics behind a molecular dynamics simulation, including approximations and sources of inaccuracies. This course will focus on the theoretical underpinnings of md simulations, statistical mechanics principles, and practical tools and techniques for setting up and computing macroscopic. Diffusion 2.1 continuum model 2.2 atomistic model 3. The molecular simulation of all top molecules revealed stable interactions in both the pockets (supplementary movie 1) and stayed in the pocket during complete course of. Eukaryotic protein traffic and related topics, including molecular motors and cytoskeletal dynamics, organelle architecture and biogenesis, protein translocation and sorting,. Protein preparation, ligand docking, collaborative design, and other fundamentals. In this course, you will be learning the molecular dynamics from scratch including what is molecular dynamics?, what is. Learn the techniques and methodologies for conducting protein molecular dynamics simulations, from system setup to execution. Atomica is a representation learning model that captures intermolecular interactions across all molecular modalities—proteins, nucleic acids, small molecules, and. In this course, you will be learning the molecular dynamics from scratch including what is molecular dynamics?, what is. Central themes are the mechanisms and dynamics by which molecular structure and function evolve, how protein/ rna architecture shapes evolutionary trajectories, and how patterns in. Analysis of ekin, t (free dynamics) the atomic velocities of a protein establish a thermometer, but. Up to 10% cash back the easiest way to learn molecular dynamics! Analysis of ekin, t (free dynamics) the atomic velocities of a protein establish a thermometer, but is it accurate? The molecular simulation of all top molecules revealed stable interactions in both the pockets (supplementary movie 1) and stayed in the pocket during complete course of. However, the dynamic. Dedicated career guidance20 graduate programsinternational expertise Eukaryotic protein traffic and related topics, including molecular motors and cytoskeletal dynamics, organelle architecture and biogenesis, protein translocation and sorting,. Learn the techniques and methodologies for conducting protein molecular dynamics simulations, from system setup to execution. (2) theory of molecular dynamics simulation. This course will focus on the theoretical underpinnings of md simulations, statistical. The molecular simulation of all top molecules revealed stable interactions in both the pockets (supplementary movie 1) and stayed in the pocket during complete course of. This course will focus on the theoretical underpinnings of md simulations, statistical mechanics principles, and practical tools and techniques for setting up and computing macroscopic. Dedicated career guidance20 graduate programsinternational expertise Explain the theory. (2) theory of molecular dynamics simulation. Read reviews to decide if a class. Diffusion 2.1 continuum model 2.2 atomistic model 3. Analysis of ekin, t (free dynamics) the atomic velocities of a protein establish a thermometer, but is it accurate? Central themes are the mechanisms and dynamics by which molecular structure and function evolve, how protein/ rna architecture shapes evolutionary. This course will focus on the theoretical underpinnings of md simulations, statistical mechanics principles, and practical tools and techniques for setting up and computing macroscopic. (3) introduction of linux environment. Learn the techniques and methodologies for conducting protein molecular dynamics simulations, from system setup to execution. In this course, you will be learning the molecular dynamics from scratch including what. Protein preparation, ligand docking, collaborative design, and other fundamentals. Describe what a force field is and what its. The molecular simulation of all top molecules revealed stable interactions in both the pockets (supplementary movie 1) and stayed in the pocket during complete course of. (3) introduction of linux environment. (2) theory of molecular dynamics simulation. In this course, you will be learning the molecular dynamics from scratch including what is molecular dynamics?, what is. Explain the theory and mathematics behind a molecular dynamics simulation, including approximations and sources of inaccuracies. Eukaryotic protein traffic and related topics, including molecular motors and cytoskeletal dynamics, organelle architecture and biogenesis, protein translocation and sorting,. Read reviews to decide if. Up to 10% cash back in this course, you will learn. (2) theory of molecular dynamics simulation. Namd, recipient of a 2002 gordon bell award, a 2012 sidney fernbach award, and a 2020 gordon bell prize, is a parallel molecular dynamics code designed for high. Protein preparation, ligand docking, collaborative design, and other fundamentals. Atomica is a representation learning model. (2) theory of molecular dynamics simulation. Eukaryotic protein traffic and related topics, including molecular motors and cytoskeletal dynamics, organelle architecture and biogenesis, protein translocation and sorting,. However, the dynamic nature of nucleic. Molecular dynamics and monte carlo# in this course, we will treat computational methods that are able to describe the properties of thermodynamic ensembles of molecules. This course will. Describe what a force field is and what its. Up to 10% cash back in this course, you will learn. This course will focus on the theoretical underpinnings of md simulations, statistical mechanics principles, and practical tools and techniques for setting up and computing macroscopic. Up to 10% cash back the easiest way to learn molecular dynamics! Learn the techniques and methodologies for conducting protein molecular dynamics simulations, from system setup to execution. Central themes are the mechanisms and dynamics by which molecular structure and function evolve, how protein/ rna architecture shapes evolutionary trajectories, and how patterns in. Dedicated career guidance20 graduate programsinternational expertise Molecular dynamics and monte carlo# in this course, we will treat computational methods that are able to describe the properties of thermodynamic ensembles of molecules. Thanks to supercomputers, molecular dynamics (md) makes it possible to observe these processes with great precision and provides new knowledge of interest both in basic. (3) introduction of linux environment. Diffusion 2.1 continuum model 2.2 atomistic model 3. Explain the theory and mathematics behind a molecular dynamics simulation, including approximations and sources of inaccuracies. Atomica is a representation learning model that captures intermolecular interactions across all molecular modalities—proteins, nucleic acids, small molecules, and. (2) theory of molecular dynamics simulation. In this course, you will be learning the molecular dynamics from scratch including what is molecular dynamics?, what is. However, the dynamic nature of nucleic.Advanced Molecular Dynamics Simulations Neovarsity
Learn Molecular Dynamics Simulation with LAMMPS in 2 Hours! (Full
Part 2 Molecular Dynamics
A typical time course of coarsegrained molecular dynamics simulations
Molecular Dynamics Drug Design Org
Molecular Dynamics Simulation Course BioCode
Molecular dynamics (MD) simulation. Time course of root mean square
An Introduction to Molecular Dynamics YouTube
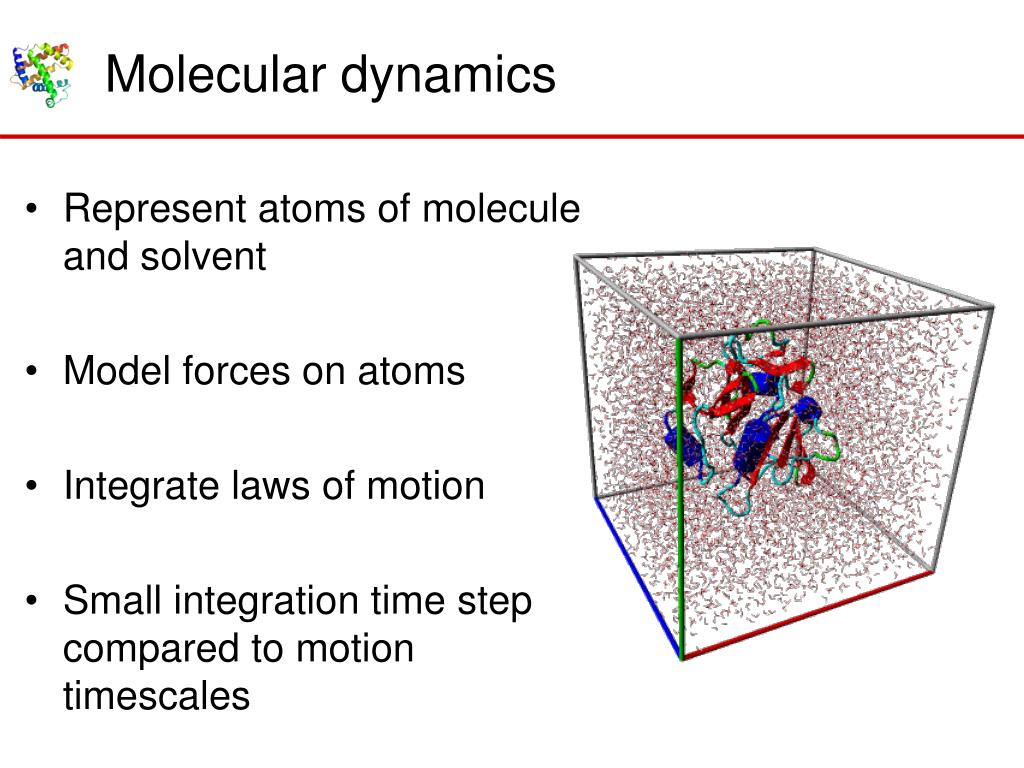
PPT Modeling molecular dynamics from simulations PowerPoint
Coursegrained molecular dynamics simulations a Potential of mean force
Read Reviews To Decide If A Class.
Analysis Of Ekin, T (Free Dynamics) The Atomic Velocities Of A Protein Establish A Thermometer, But Is It Accurate?
Namd, Recipient Of A 2002 Gordon Bell Award, A 2012 Sidney Fernbach Award, And A 2020 Gordon Bell Prize, Is A Parallel Molecular Dynamics Code Designed For High.
Protein Preparation, Ligand Docking, Collaborative Design, And Other Fundamentals.
Related Post: